Research attention has been powered to understand the functional roles of non-coding RNAs (ncRNAs). Many studies have demonstrated their deregulation in cancer and other human disorders. ncRNAs are also present in extracellular human body fluids such as serum and plasma, giving them a great potential as non-invasive biomarkers. However, non-coding RNAs have been relatively recently discovered and a comprehensive database including all of them is still missing. Reconstructing and visualizing the network of ncRNAs interactions are important steps to understand their regulatory mechanism in complex systems. This work presents ncRNA-DB, a NoSQL database that integrates ncRNAs data interactions from a large number of well established on-line repositories. The interactions involve RNA, DNA, proteins, and diseases. ncRNA-DB is available at http://ncrnadb.scienze.univr.it/ncrnadb/. It is equipped with three interfaces: web based, command-line, and a Cytoscape app called ncINetView. By accessing only one resource, users can search for ncRNAs and their interactions, build a network annotated with all known ncRNAs and associated diseases, and use all visual and mining features available in Cytoscape.
1. INTRODUCTION
After the sequencing of the human genome, it became evident that only 20,000 genes are protein-coding, while over 98% of all genes are untranslated non-protein-coding RNAs (ncRNAs) (ENCODE Project Consortium, 2012). During the last years, thousands of ncRNAs have been identified in the eukaryotic transcriptome (Khalil et al., 2009; Bu et al., 2011). Usually, ncRNAs are divided into two groups according to their length: short ncRNAs, consisting of
4. CONCLUSION
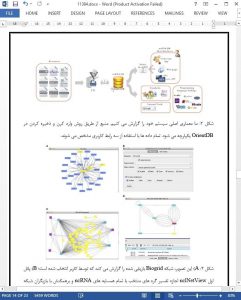
In this paper, we have presented ncRNA-DB, an integrated database storing knowledge concerning ncRNAs, genes, and associated diseases. The system has been implemented within the NoSQL database OrientDB. It stores data coming from several leading resources such as HGNC, lncRNAdb, circ2Traits, HMDD, lncRNADiseases, miRandola, miRTarBase, NON-CODE, and NPInter. ncRNA-DB can be queried trough three interfaces. A Cytoscape App, named ncINetView, allows to annotate biological networks with ncRNA knowledge. A web app and a command-line interface, which allows users to query the ncRNA-DB and to extract information in a text format. The aim of the proposed system is to give a comprehensive access to all the knowledge available in the literature concerning ncRNAs and associated diseases. As a key characteristics, the integrated data aim to reduce the problem of different nomenclatures used by different sources.