Abstract
A translational regulatory mechanism for Tomato golden mosaic virus (TGMV) complementary-sense gene expression has been characterized. TGMV transcribes two mRNAs, AL-1935 and AL-1629 transcripts, both of which contain the AL2 and AL3 open reading frames. However, AL2 is only expressed from AL-1629 whereas AL3 is expressed from both. Three AUG translation initiation codons are located upstream of both the AL2 and AL3 coding regions, within the 5′-untranslated region (UTR) of the AL-1935 transcript. Translation can initiate at the first AUG, specifying the C-terminal 122 amino acids of the AL1 protein (cAL1). Initiation of translation at this AUG is inhibitory for the downstream expression of both AL2 and AL3. This is most likely due to the terminator codon of cAL1 being positioned after the AUG initiation codon for the AL2 ORF. The mechanism by which AL3 is expressed from AL-1935 is currently unknown but a gap between the cAL1 termination codon and the start of AL3 suggests that it may involve reinitiation and/or internal initiation. In contrast, expression of AL3 from AL-1629 most likely occurs via leaky ribosome scanning since the AL3 initiation codon occurs before the terminator codon of AL2. Mutation of the AUG encoding cAL1 in the curtovirus, Spinach curly top virus, leads to increased infectivity as measured by a shorter latent period. Together this suggests that geminiviruses use a post-translational regulatory mechanism to regulate the synthesis of viral proteins important for replication and suppression of host defenses.
Introduction
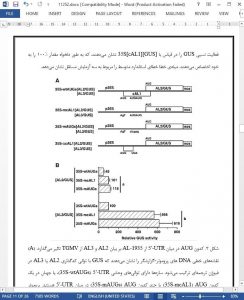
The bipartite genome of Tomato golden mosaic virus (TGMV), a begomovirus, consists of two DNA components, A and B, of approximately 2.6 kb (Hamilton et al., 1984). Replication of the single stranded DNA genome occurs in the nuclei of infected plant cells producing double-stranded (ds) DNA replicative form (RF) intermediates (Stenger et al., 1991). Virus-specific mRNAs are transcribed from RF DNA in a bidirectional manner from an approximately 230 nt region, common to both DNA components (Fig. 1A) (Fontes et al., 1994; Hanley-Bowdoin et al., 1990; Petty et al., 1988; Sunter and Bisaro, 1989; Sunter et al., 1989, 1993). Viral RNAs are polyadenylated and initiate downstream of either consensus TATA box or initiator elements indicative of transcription mediated by host RNA polymerase II. It is unknown at this time whether TGMV transcripts are capped at the 5′-end. A single virion-sense RNA is transcribed from TGMV DNA A initiating at nt 319 (AR-319), providing the template for translation of CP (Sunter et al., 1989). Complementary-sense transcription is more complex, producing multiple overlapping RNAs with different 5′-ends but a common 3′-end. The largest transcript initiates at nt 62 (AL-62), encodes the entire left side of TGMV A (Sunter et al., 1989), and is the only transcript that can encode full-length AL1 protein. However, AL-62 also has the potential to code for the AL2, AL3 and AL4 proteins.
Plant inoculation
Healthy N. benthamiana, Arabidopsis or spinach (Spinacea oleracea, var. viroflay) were inoculated with Agrobacterium cultures containing either tandemly repeated copies of wild type or mutant SCTV or TGMV DNA, using a standard dose (O.D.600 = 1.0) as described previously (Baliji et al. 2004). Inoculated plants were scored for the appearance of symptoms typical of an SCTV or TGMV infection on systemically infected tissue on a daily basis. Latent periods and ID50 measurements were made as described previously (Sunter et al., 2001). DNA was isolated from infected, and healthy mock-inoculated plants and analyzed for viral DNA accumulation as described previously (Baliji et al., 2007).